Research Interests
Our Interest
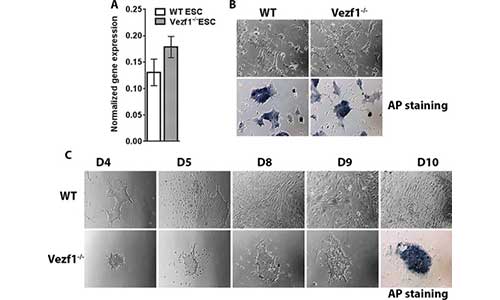
Gowher lab has an ongoing interest in elucidating the mechanism of insulation by the insulator-binding protein Vezf1. Previous studies showed that Vezf1 regulates the expression and splicing of Dnmt3b in mouse ES cells. Our recent data strongly indicates its role in the differentiation of endothelial cells and cardiomyocyte progenitors. ES cell differentiation into various cells cell types is a powerful tool that will be used to elucidate mechanisms by which distinct transcriptional programs are regulated by chromatin insulation and long-range contacts.

Area of Expertise: Regulation of DNA methylation in development and disease
Recent Findings
Recent findings from our lab elucidated mechanisms that regulate target site recognition and catalytic activity of DNMT3A and 3B in the normal and diseased states. For example, we showed that in normal embryonic stem cells (ESC), histone demethylation by Lsd1 guides DNMT3A activity (1), whereas, an aberrant Oct4 expression in embryonal carcinoma cells (ECC) inhibits Lsd1 activity and downstream DNMT3A activity (2). Similarly, in normal cells, the cooperative catalytic mechanism, which supports the high catalytic turnover and DNA sequence preference of DNMT3A (3), is disrupted by DNMT3A R882H mutation in AML cancer cells and the variant enzyme gains DNMT3B-like properties (4).
Embryonic stem cell differentiation
Using embryonic stem cell differentiation as a developmental model system, we ask the following questions:
How is the target site specificity of DNMT3 enzymes guided by their unique structural and enzymatic properties?
How is the target site specificity of DNMT3 enzymes guided by chromatin and non-chromatin interaction partners?
How are the tissue-specific expression and alternative splicing of DNA MTases regulated?
What is the contribution of the major and minor isoforms of DNMT3B in the regulation of DNA methylation?
These questions are addressed by designing experiments that elucidate mechanisms facilitating communication between various epigenetic modulators such as histone modifications, non-coding RNA, and DNA methylation and how the combinatorial output of epigenetic signals impacts the transcriptional machinery.

